Senior Investigator Research Interests
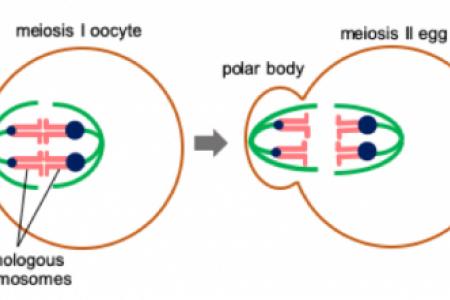
One of the fundamental rules in biology is Mendel’s Law of Segregation, which states that two alleles of a gene have an equal chance to be transmitted to gametes. Such segregation will result in the typical 3:1 phenotype ratio as in Mendel’s experiments with garden peas. However, this law can be violated by selfish genetic elements, which exploit meiosis to increase their own rate of transmission. This genetic cheating in meiosis, meiotic drive, potentially occurs whenever haploid gametes are produced from diploid parents, a process inherent to all sexually reproducing eukaryotes. Female meiosis provides an opportunity for genetic elements to cheat because of the highly asymmetric cell division: only chromosomes segregated to the egg are transmitted to the descendants, while the rest are degraded in polar bodies (Fig. 1). Meiotic drive impacts genetics, evolution, and fertility, as selfish elements distort transmission ratios of linked genes and allele frequencies in populations and manipulate gamete production. Also, meiotic drive is typically associated with fitness costs to individuals that carry selfish elements. Using mouse oocytes as a model, the laboratory of Chromosome Dynamics and Evolution, led by Dr. Takashi Akera focuses on both the cell biological basis and evolutionary consequences of meiotic drive. Elucidating how selfish elements exploit the asymmetry in female meiosis will also provide a deep understanding of female meiosis itself, which is error-prone, causing fertility issues in humans. Therefore, the lab also studies basic chromosome segregation “rules” in oocytes, which can be exploited by selfish elements.
Centromeres direct chromosome segregation, and therefore are the genetic elements with the best opportunity to cheat the segregation process. Meiotic drive of selfish centromeres, or centromere drive, can explain the “centromere paradox”: rapid evolution of both centromere DNA sequences and genes encoding centromere-binding proteins despite conserved centromere function in segregation. The centromere drive theory is based on the idea that natural selection favors centromere DNA sequences that act selfishly in female meiosis. Fitness costs associated with centromere drive would also select for alleles of centromere-binding proteins that suppress centromere drive. Thus, centromere DNA and centromere proteins continually evolve in conflict with each other, analogous to a molecular arms race between viruses and the immune system. This theory has been influential, but it was largely unknown how centromere drive happens cell biologically.
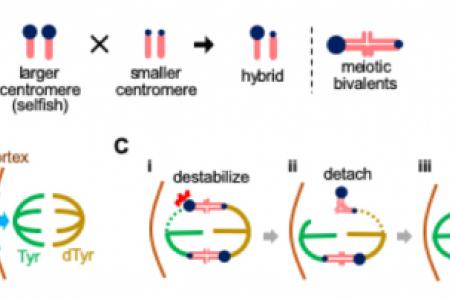
The lab has established the first experimental system for centromere drive in mice, leading to three major advances. First, we showed that centromeres with expanded satellite repeats act selfishly to preferentially orient towards the egg pole of the meiotic spindle to remain in the egg (Fig. 1 and 2A). Second, we found that CDC42 GTPase signaling from the cell cortex regulates microtubules (MTs) to induce asymmetry in MT tyrosination within the spindle and that non-Mendelian segregation depends on this asymmetry (Fig. 2B). Third, we demonstrated that high MT-destabilizing activity confers selfishness to centromeres. Centromeres incorporate both MT-binding and counteracting MT-destabilizing activities: the former attaches chromosomes to the spindle, and the latter promotes re-orientation of incorrect attachments to prevent segregation errors. Selfish centromeres exploit the same destabilizing activity, by selectively promoting re-orientation to bias their segregation to the egg (Fig. 2C). Together, these findings provided first insights into how selfish mouse centromeres exploit the inherent asymmetry in female meiosis to challenge Mendel.
Centromere drive and the resulting conflicts between centromere DNA and centromere proteins can generate distinct evolutionary trajectories in different populations, explaining the large divergence in centromere DNA even between closely-related species or strains. This divergence in centromeres could be a driving force of speciation by causing hybrid incompatibility in essential centromere functions. A current focus of the Akera lab is to take advantage of the rich natural variation and species divergence in mice, which has not been so much exploited in biomedical research, to reveal the impact of centromere evolution on reproductive isolation.
Because centromere-MT interactions have been extensively studied, models for how centromeres can cheat to bias their segregation are relatively intuitive. However, other driving loci reside outside centromeres, and mechanisms for their non-Mendelian segregation and causes of their fitness costs are completely unknown, except for one example in maize. Another major focus of the lab is to address the cell biological mechanisms of meiotic drive of non-centromeric loci and the associated fitness costs in animals. We are currently working on the selfish R2d2 locus in mice.
Meiotic drive is fundamental to sexual reproduction and has been recognized as a powerful force in genetics and evolutionary biology since first described in maize in 1942. The underlying mechanisms have long been mysterious to cell biologists. The Akera lab tackles this exciting problem, all the way from developing experimental systems to revealing how selfish elements challenge Mendel and affect fitness. Moreover, our work will lead to a deeper understanding of the interactions between chromosomes and spindle MTs. These interactions are highly error-prone in humans and a major cause of infertility, which could be caused by selfish behaviors of meiotic drivers.
Watch a rigged game of tug-of-war inside the egg cells of mammals.
Meet the Team

Takashi Akera, Ph.D.
Dr. Takashi Akera graduated from the University of Tokyo with a Ph.D. in Biophysics and Biochemistry in 2014. He conducted postdoctoral research at the University of Pennsylvania in the laboratory of Dr. Michael Lampson from 2015 to 2019. He received both Holtzer Award for outstanding postdoctoral research in Cell and Developmental Biology and Kaushal Award for excellence in postdoctoral research in Genetics from the University of Pennsylvania in 2018. He was also a finalist for the ASCB Porter Prize for Research Excellence Award in 2018. Dr. Akera joined the NHLBI in 2019 as an Earl Stadtman tenure-track Investigator and is a member of the American Society for Cell Biology.
Contact the lab

Betsy Clark, PhD

Warif El Yakoubi, PhD

Eddie (Bo) Pan, PhD

Zaak Walton

Takaya Totsuka, Ph.D.

Duílio Silva, PhD

Sebastian Khan